December 2022
nature genetics
Spurious transcription causing innate immune responses is prevented by 5-hydroxymethylcytosine
Fan Wu, Xiang Li, Mario Looso, Hang Liu, Dong Ding, Stefan Günther, Carsten Kuenne, Shuya Liu, Norbert Weissmann, Thomas Boettger, Ann Atzberger, Saeed Kolahian, Harald Renz, Stefan Offermanns, Ulrich Gärtner, Michael Potente, Yonggang Zhou, Xuejun Yuan & Thomas Braun
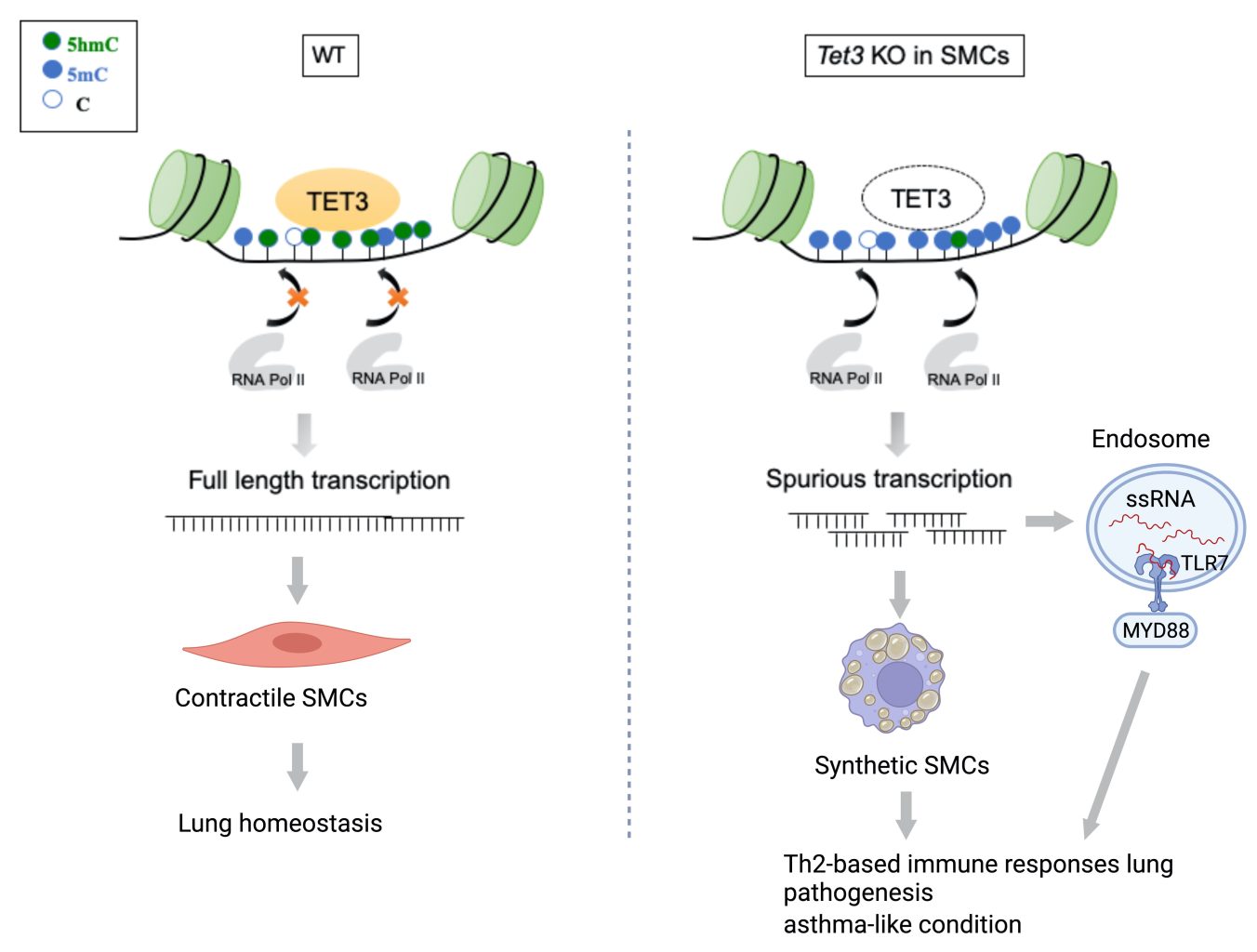
The dynamic DNA methylation/demethylation on cytosine is a major epigenetic mechanism to regulate smooth muscle cell (SMC) plasticity that is substantially deregulated under pathophysiological conditions. In this study, we examined the function of Tet3, a DNA demethylases that convert 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC), in controlling lung SMC and tissue homeostasis. Tet3 ablation in SMC evokes massive inflammation, airway remodeling and phenotypical shift of SMC from contractile to more secretory synthetic state. Integrative analysis of Omics data of RNA-seq, ChIP-seq and MeDIP-seq reveals that 5hmC is largely enriched in gene bodies of highly expressed genes like contractile genes. Tet3 inactivation leads to substantial reduction of 5hmC level, intragenic entry of RNA polymerase II pSer5 and subsequent spurious transcription initiation within highly transcribed genes. Ultimately, accumulated aberrant RNA transcripts activate endosomal TLR signaling pathway and provoke cytokine/chemokine production. Taken together, our results demonstrate a novel epigenetic mechanism that link DNA demethylation, spurious transcription and innate immune response during lung pathogenesis.